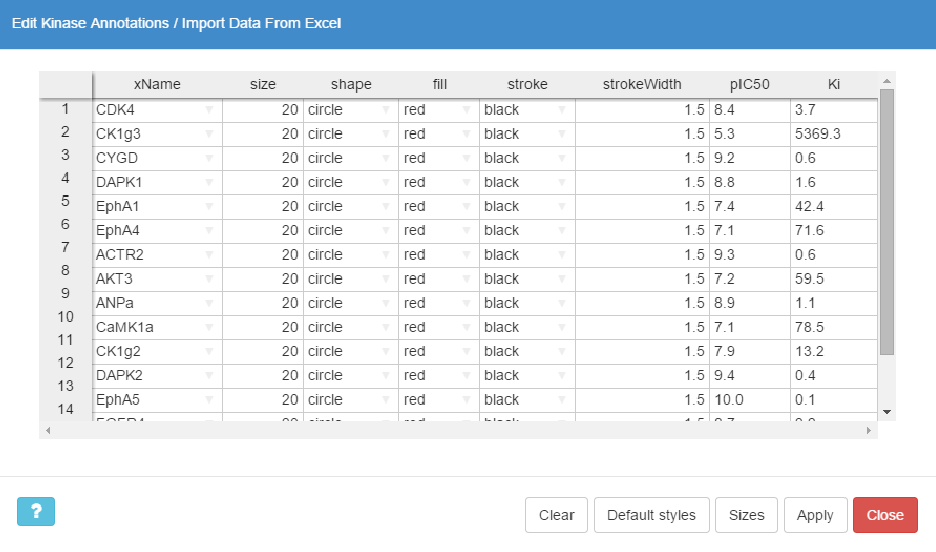
This button opens up an interactive spreadsheet editor that shows the currently rendered annotations. Here you can add or delete kinase annotations, and modify their styles, colors and sizes in a convenient manner.
To add a new annotation you just need to provide a valid kinase name in the xName column or select it from the drop-down list. When style definitions are missing, the default style (red circle, 20px in diameter) will be automatically used for rendering. Alternatively, you can fill the missing styles by clicking the Default styles button.
You can directly paste data into KinMap spreadsheet from other editors, e.g. MS Excel . In such case, at least the first column is required (a valid kinase name). If you would like to paste style attributes, then the columns must be in the right order.
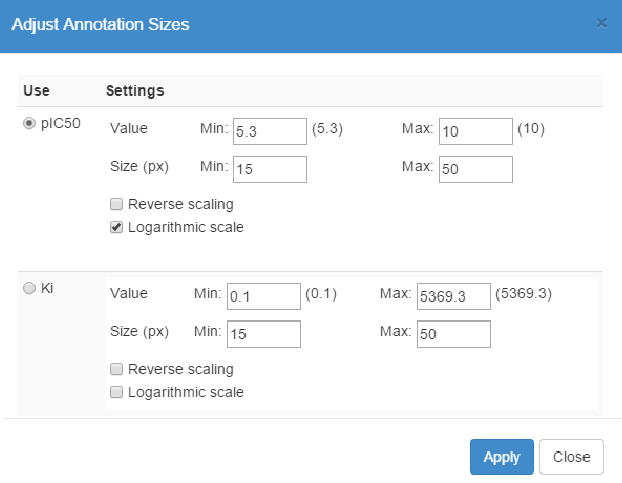
Click on the Sizes button to open the Adjust Annotation Sizes panel. Here, you can define the sizes of kinase annotations using the values in one column from the input, e.g. biochemical data imported from a CSV file (see below).
You can import annotations directly from text files, comma-separated values files, or KMAP files to KinMap (see Supported Formats). At minimum, KinMap expects the input to contain the names of kinases to be annotated, i.e. style definitions are optional, as the missing ones will be automatically filled with the default style.
It's important to note that KinMap can read any arbitrary data in the input, e.g. biochemical assay results, selectivity scores, etc. The additional data can be used, for instance, to adjust annotation sizes or as sources of information for the interactive kinome view.
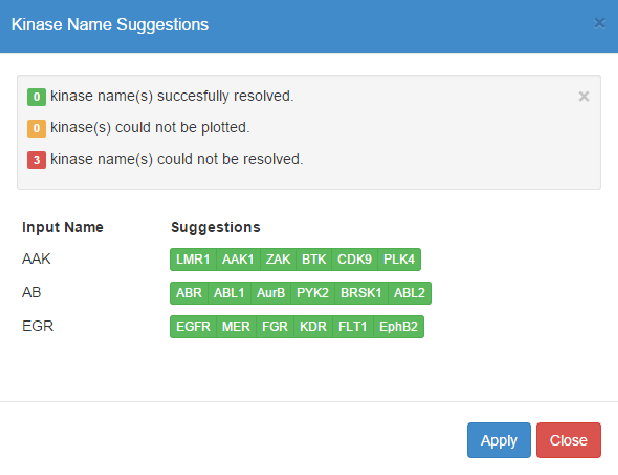
KinMap employs a unified KinMap name (xName) to define each kinase annotation, and it allows multiple annotations to be associated with the same kinase node on the tree. However, KinMap accepts other valid names or abbreviations in the input. The built-in parsing function can recognize names used by Manning et al., recommended by the UniProt Consortium, approved by the HUGO Gene Nomenclature Committee; as well as any of the alternative names listed in these resources.
When kinase name(s) in the input cannot be uniquely resolved to a valid xName, e.g. incomplete or ambiguous names, the Validate Names panel allows you choose the intended kinase(s) from a prioritized list of potential matches.
KinMap keeps track of rendered kinase annotations and any changes made to them, e.g. readjusting sizes or modifying colors. You can revert to any of the previous annotations using the History drop-down list.
Use the Zoom/Pan tools to adjust the viewport of the kinome tree.
Show/Hide current kinase annotations. Note that the hidden annotations are not deleted.
Show/Hide the tree of the atypical kinases (bottom-left) along with its annotations.
Activate/Deactivate the Interactive Kinome View mode. In this mode, small yellow square controls are shown above each kinase in the tree. Hovering with the mouse over these controls opens a floating widget that lists key information of the respective kinase.
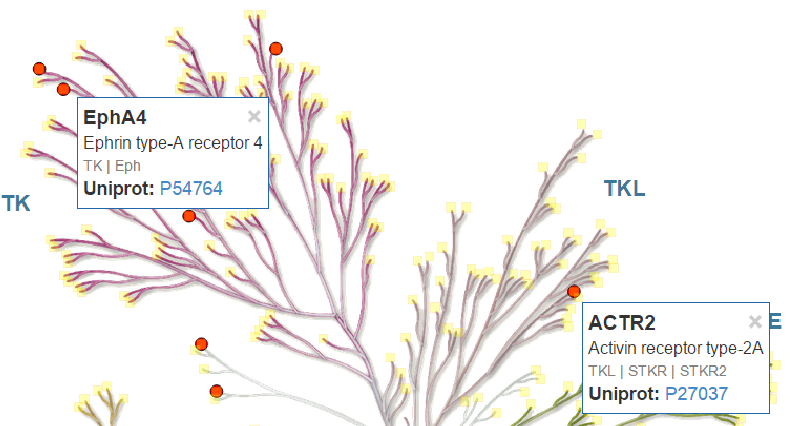
Clicking the square controls makes the floating widget persistent. You can dismiss the floating widget by clicking the × in the top-right corner. Alternatively, you can dismiss all widgets by selecting Delete > Close All Info Box(es). The information shown in the floating widget can be key data of the respective kinase or additional data imported from a CSV file. You can customize this from the Hover Info Settings panel.
Deletes all current annotations and append a respective record in the annotation history.
Delete the crosshair highlighting the kinase corresponding to the result of the latest query in the search box at the top of the page.
Close all the floating information widgets from the Interactive Kinome View.
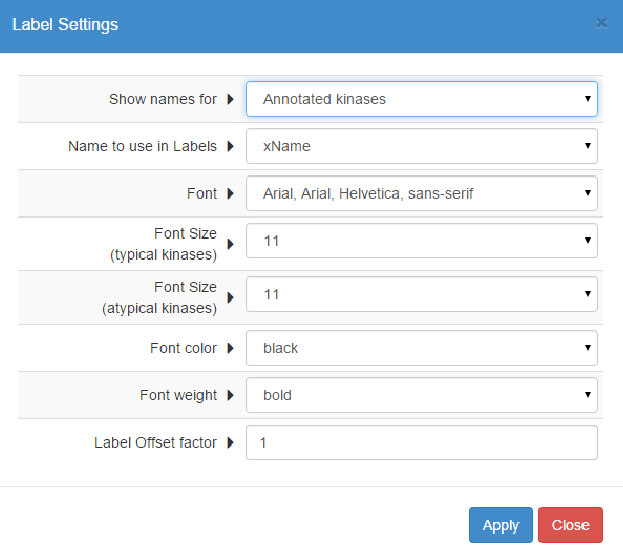
Here you can customize the subset of kinases to show labels for, and customize the style of these labels (font style, size, color and weight).
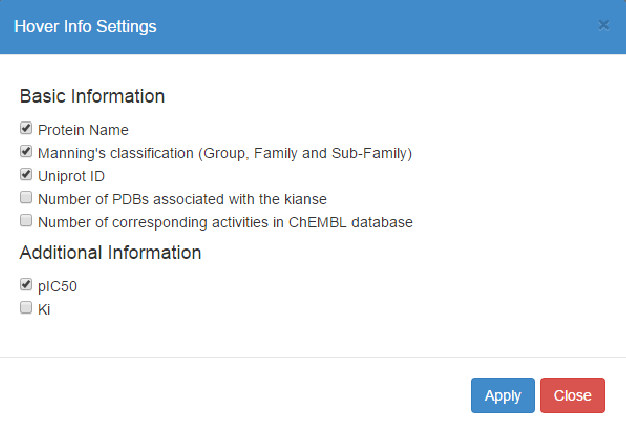
Here you can customize the information displayed in the widgets in the Interactive Kinome View. KinMap provides some basic kinase information (name, classification, counts of available experimental data), but you can also show additional information from imported input files.
You can download the annotated kinome tree as SVG image or PNG image (normal or high-resolution). In addition, you can export the kinome tree annotations to an editable format for future use.
The minimalistic plain text format saves flat lists of kinases and concise style directives making it suitable for simple annotations showing subgroups of kinases. However, this format does not preserve non-style information such as assay results.
The CSV format is more expansive, preserving detailed style information and additional data defined by the user. Moreover, the CSV format can be viewed and edited using popular spreadsheet software, e.g. Microsoft Excel and OpenOffice Calc. Hence, the CSV format is the most practical for exchange of kinome tree annotations.
Contains the same data as the CSV format, but can be used only for export. Due to XML tag name limitations, non-alphanumeric charachters in column names are replaced with an underscore.
The KMAP format is a non-proprietary format based on the JSON standard. It contains additional metadata necessary to exactly reproduce the view of KinMap, e.g. text label size and color settings, zoom level, interactive mode settings.
KinMap can handle up to 2000 kinase annotations on common desktop computers and laptops without any degradation in performance. In some extreme practice runs, KinMap was tested with nearly 10’000 annotations and was still functioning properly with only a small rendering lag of a few seconds.